MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media:

MLZ (eng)
Lichtenbergstr.1
85748 Garching
08.01.2018
A new “neutron window” onto membrane protein structures
Membrane proteins represent an important class of bio-macromolecules performing a diverse set of biological roles. They form, for example, gates to import nutrients and chemical messengers inside the cell or for the release of waste products. Furthermore, they are often targeted by active ingredients of pharmaceuticals. Although knowledge of their structure is of paramount importance for a better understanding of a multitude of biological processes, they are – in comparison to soluble globular proteins – heavily underrepresented in the major worldwide database for three-dimensional structural data of large biological molecules. This is mainly due to difficulties in the expression, crystallization and solubilization of membrane proteins that hinder the application of established experimental techniques such as crystallography and nuclear magnetic resonance.
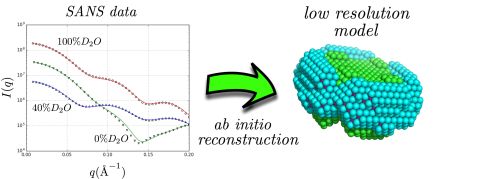
Neutron scattering curves of complexes of the membrane protein aquaporin-0 and the detergent n-Dodecyl β-D-Maltoside at various solvent contrasts (left), result of the ab-initio low resolution shape reconstruction using the discussed methodology (right). © Forschungszentrum Jülich
During the past 15 years, small-angle scattering of neutrons (SANS) and X-rays (SAXS) have become increasingly popular in the structural biology community as complementary tools for low-resolution (1-2nm) structure determination of proteins and nucleic acids in solution. However, due to the need for detergents or other molecular assemblies such as bicelles, nanodiscs or amphipols for the solubilization of membrane proteins, the extraction of the scattering signal belonging to membrane proteins is not trivial, as not only the proteins but also the detergents or bicelles etc. contribute to the scattering. Furthermore, there is a relative lack of theoretical and computational tools for the analysis of scattering data coming from protein/detergent complexes.
In a recent paper published in the Biophysical Journal, neutron scattering experts from Forschungszentrum Jülich illustrate that small-angle neutron scattering data obtained at different solvent contrasts (i.e. different D2O/H2O ratios) permit, through the use of an appropriate model and minimization procedure, the reconstruction of the low resolution structure of the different parts of detergent/protein complexes. This model attempts to fit the experimental data using a multiphase coarse-grained approach that encompasses a set of physical constraints adapted to the anticipated assembly of detergents around the protein surface. A related software program named “DANVILLE” was developed and is provided freely to the academic community.
“The methodology presented here coupled with high quality data acquisition at current and future advanced neutron sources provides a powerful new approach for attacking the notoriously difficult problem of membrane protein structure determination, especially in the lower molecular weight window below 80-100 kDa that falls outside the current range of cryo-electron microscopy investigations”, argues Dr. Alexandros Koutsioumpas, instrument scientist at the Jülich Centre for Neutron Science (JCNS) at the Heinz Maier-Leibnitz Zentrum (MLZ).
Text: A. Wenzik / JCNS
Original publication:
A. Koutsioubas
Low Resolution Structure Determination of Detergent Solubilised Membrane Proteins from Small Angle Scattering Data
Biophys.J. 2017;113(11):2373-2382, DOI: 10.1016/j.bpj.2017.10.003
Available via Open Access until the end of january 2018
MLZ is a cooperation between:
 > Technische Universität München
> Technische Universität München > Helmholtz-Zentrum Hereon
> Helmholtz-Zentrum Hereon
 > Forschungszentrum Jülich
> Forschungszentrum Jülich
MLZ is a member of:
 > LENS
> LENS > ERF-AISBL
> ERF-AISBL
MLZ on social media:



